-

David Murrugarra
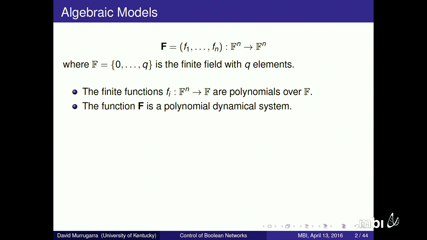
Understanding the regulatory mechanisms in molecular interaction networks is an important goal in systems biology. This talk will focus on processes at the molecular level that determine the state of an individual cell, involving signaling or cell regulation. The mathematical framework to be used is that of Boolean networks and their multi-state generalization. These models represent the interactions of different molecular species as logical rules that describe how these species combine to regulate others. Regulatory rules that appear in published models tend to have special features such as the property of being nested canalizing, a concept inspired by the concept of canalization in evolutionary biology. This talk will survey a set of results about nested canalizing rules and how these constrain network dynamics. It has been shown that networks comprised of nested canalizing functions have dynamic properties that make them suitable for modeling gene regulatory networks, namely small number of attractors and short limit cycles. In this talk, I will discuss a normal form as polynomial function that applies to any Boolean or multi-state function. This description provides a partition of the inputs of any Boolean function or multi-state function into canalizing and non-canalizing variables and, within the canalizing ones, we can categorize the input variables into layers of canalization. I will also describe the structure of the non-canalizing variables. Applications for how to use this normal form and some other properties of these functions will be given at the end of the talk.
-
David Murrugarra
This talk will introduce a new modeling framework for gene regulatory networks that incorporates state dependent delays and that is able to capture the cell-to-cell variability. This framework will be presented in the context of finite dynamical systems, where each gene can take on a finite number of states, and where time is also a discrete variable. The state dependent delays represent the time delays of activation and degradation. One of the new features of this framework is that it allows a finer analysis of discrete models and the possibility to simulate cell populations. Applications presented will use one of the best known stochastic regulatory networks, that is involved in controlling the outcome of lambda phage infection of bacteria.
-

David Murrugarra
Boolean networks are an important class of computational models for molecular interaction networks. Boolean canalization, a type of hierarchical clustering of the inputs of a Boolean function, has been extensively studied in the context of network modeling where each layer of canalization adds a degree of stability in the dynamics of the network. Recently, dynamic network control approaches have been used for the design of new therapeutic interventions and for other applications such as stem cell reprogramming. This talk will discuss the role of canalization in the control of Boolean molecular networks. A method for identifying the potential control edges in the wiring diagram of a network for avoiding undesirable state transitions will be presented. The method is based on identifying appropriate input-output combinations on undesirable transitions that can be modified using the edges in the wiring diagram of the network. Moreover, a method for estimating the number of changed transitions in the state space of the system as a result of an edge deletion in the wiring diagram will be presented. These two complementary methods can help in the selection of appropriate controllers such as for minimizing the side effects resulting from an edge manipulation.
-
David Murrugarra
Many problems in biomedicine and other areas of the life sciences can be characterized as control problems, with the goal of finding strategies to change a disease or otherwise undesirable state of a biological system into another, more desirable, state through an intervention, such as a drug or other therapeutic treatment. The identification of such strategies is typically based on a mathematical model of the process to be altered through targeted control inputs. This talk focuses on processes at the molecular level that determine the state of an individual cell, involving signaling or gene regulation. The mathematical model type considered is that of Boolean networks. The potential control targets can be represented by a set of nodes and edges that can be manipulated to produce a desired effect on the system. This talk presents a method for the identification of potential intervention targets in Boolean molecular network models using algebraic techniques. The approach exploits an algebraic representation of Boolean networks to encode the control candidates in the network wiring diagram as the solutions of a system of polynomials equations, and then uses computational algebra techniques to find such controllers. Additionally, a formula, based on the properties of Boolean canalization, for estimating the number of changed transitions in the state space of the system as a result of an edge deletion in the wiring diagram will be discussed. Finally, an optimal control algorithm for the identification of the best combination of control actions in a stochastic system will be presented.
 David MurrugarraUnderstanding the regulatory mechanisms in molecular interaction networks is an important goal in systems biology. This talk will focus on processes at the molecular level that determine the state of an individual cell, involving signaling or cell regulation. The mathematical framework to be used is that of Boolean networks and their multi-state generalization. These models represent the interactions of different molecular species as logical rules that describe how these species combine to regulate others. Regulatory rules that appear in published models tend to have special features such as the property of being nested canalizing, a concept inspired by the concept of canalization in evolutionary biology. This talk will survey a set of results about nested canalizing rules and how these constrain network dynamics. It has been shown that networks comprised of nested canalizing functions have dynamic properties that make them suitable for modeling gene regulatory networks, namely small number of attractors and short limit cycles. In this talk, I will discuss a normal form as polynomial function that applies to any Boolean or multi-state function. This description provides a partition of the inputs of any Boolean function or multi-state function into canalizing and non-canalizing variables and, within the canalizing ones, we can categorize the input variables into layers of canalization. I will also describe the structure of the non-canalizing variables. Applications for how to use this normal form and some other properties of these functions will be given at the end of the talk.
David MurrugarraUnderstanding the regulatory mechanisms in molecular interaction networks is an important goal in systems biology. This talk will focus on processes at the molecular level that determine the state of an individual cell, involving signaling or cell regulation. The mathematical framework to be used is that of Boolean networks and their multi-state generalization. These models represent the interactions of different molecular species as logical rules that describe how these species combine to regulate others. Regulatory rules that appear in published models tend to have special features such as the property of being nested canalizing, a concept inspired by the concept of canalization in evolutionary biology. This talk will survey a set of results about nested canalizing rules and how these constrain network dynamics. It has been shown that networks comprised of nested canalizing functions have dynamic properties that make them suitable for modeling gene regulatory networks, namely small number of attractors and short limit cycles. In this talk, I will discuss a normal form as polynomial function that applies to any Boolean or multi-state function. This description provides a partition of the inputs of any Boolean function or multi-state function into canalizing and non-canalizing variables and, within the canalizing ones, we can categorize the input variables into layers of canalization. I will also describe the structure of the non-canalizing variables. Applications for how to use this normal form and some other properties of these functions will be given at the end of the talk. David MurrugarraThis talk will introduce a new modeling framework for gene regulatory networks that incorporates state dependent delays and that is able to capture the cell-to-cell variability. This framework will be presented in the context of finite dynamical systems, where each gene can take on a finite number of states, and where time is also a discrete variable. The state dependent delays represent the time delays of activation and degradation. One of the new features of this framework is that it allows a finer analysis of discrete models and the possibility to simulate cell populations. Applications presented will use one of the best known stochastic regulatory networks, that is involved in controlling the outcome of lambda phage infection of bacteria.
David MurrugarraThis talk will introduce a new modeling framework for gene regulatory networks that incorporates state dependent delays and that is able to capture the cell-to-cell variability. This framework will be presented in the context of finite dynamical systems, where each gene can take on a finite number of states, and where time is also a discrete variable. The state dependent delays represent the time delays of activation and degradation. One of the new features of this framework is that it allows a finer analysis of discrete models and the possibility to simulate cell populations. Applications presented will use one of the best known stochastic regulatory networks, that is involved in controlling the outcome of lambda phage infection of bacteria. David MurrugarraBoolean networks are an important class of computational models for molecular interaction networks. Boolean canalization, a type of hierarchical clustering of the inputs of a Boolean function, has been extensively studied in the context of network modeling where each layer of canalization adds a degree of stability in the dynamics of the network. Recently, dynamic network control approaches have been used for the design of new therapeutic interventions and for other applications such as stem cell reprogramming. This talk will discuss the role of canalization in the control of Boolean molecular networks. A method for identifying the potential control edges in the wiring diagram of a network for avoiding undesirable state transitions will be presented. The method is based on identifying appropriate input-output combinations on undesirable transitions that can be modified using the edges in the wiring diagram of the network. Moreover, a method for estimating the number of changed transitions in the state space of the system as a result of an edge deletion in the wiring diagram will be presented. These two complementary methods can help in the selection of appropriate controllers such as for minimizing the side effects resulting from an edge manipulation.
David MurrugarraBoolean networks are an important class of computational models for molecular interaction networks. Boolean canalization, a type of hierarchical clustering of the inputs of a Boolean function, has been extensively studied in the context of network modeling where each layer of canalization adds a degree of stability in the dynamics of the network. Recently, dynamic network control approaches have been used for the design of new therapeutic interventions and for other applications such as stem cell reprogramming. This talk will discuss the role of canalization in the control of Boolean molecular networks. A method for identifying the potential control edges in the wiring diagram of a network for avoiding undesirable state transitions will be presented. The method is based on identifying appropriate input-output combinations on undesirable transitions that can be modified using the edges in the wiring diagram of the network. Moreover, a method for estimating the number of changed transitions in the state space of the system as a result of an edge deletion in the wiring diagram will be presented. These two complementary methods can help in the selection of appropriate controllers such as for minimizing the side effects resulting from an edge manipulation. David MurrugarraMany problems in biomedicine and other areas of the life sciences can be characterized as control problems, with the goal of finding strategies to change a disease or otherwise undesirable state of a biological system into another, more desirable, state through an intervention, such as a drug or other therapeutic treatment. The identification of such strategies is typically based on a mathematical model of the process to be altered through targeted control inputs. This talk focuses on processes at the molecular level that determine the state of an individual cell, involving signaling or gene regulation. The mathematical model type considered is that of Boolean networks. The potential control targets can be represented by a set of nodes and edges that can be manipulated to produce a desired effect on the system. This talk presents a method for the identification of potential intervention targets in Boolean molecular network models using algebraic techniques. The approach exploits an algebraic representation of Boolean networks to encode the control candidates in the network wiring diagram as the solutions of a system of polynomials equations, and then uses computational algebra techniques to find such controllers. Additionally, a formula, based on the properties of Boolean canalization, for estimating the number of changed transitions in the state space of the system as a result of an edge deletion in the wiring diagram will be discussed. Finally, an optimal control algorithm for the identification of the best combination of control actions in a stochastic system will be presented.
David MurrugarraMany problems in biomedicine and other areas of the life sciences can be characterized as control problems, with the goal of finding strategies to change a disease or otherwise undesirable state of a biological system into another, more desirable, state through an intervention, such as a drug or other therapeutic treatment. The identification of such strategies is typically based on a mathematical model of the process to be altered through targeted control inputs. This talk focuses on processes at the molecular level that determine the state of an individual cell, involving signaling or gene regulation. The mathematical model type considered is that of Boolean networks. The potential control targets can be represented by a set of nodes and edges that can be manipulated to produce a desired effect on the system. This talk presents a method for the identification of potential intervention targets in Boolean molecular network models using algebraic techniques. The approach exploits an algebraic representation of Boolean networks to encode the control candidates in the network wiring diagram as the solutions of a system of polynomials equations, and then uses computational algebra techniques to find such controllers. Additionally, a formula, based on the properties of Boolean canalization, for estimating the number of changed transitions in the state space of the system as a result of an edge deletion in the wiring diagram will be discussed. Finally, an optimal control algorithm for the identification of the best combination of control actions in a stochastic system will be presented.